Artificial Intelligence and Computational Approaches for Epilepsy
Article information
Abstract
Studies on treatment of epilepsy have been actively conducted in multiple avenues, but there are limitations in improving its efficacy due to between-subject variability in which treatment outcomes vary from patient to patient. Accordingly, there is a growing interest in precision medicine that provides accurate diagnosis for seizure types and optimal treatment for an individual epilepsy patient. Among these approaches, computational studies making this feasible are rapidly progressing in particular and have been widely applied in epilepsy. These computational studies are being conducted in two main streams: 1) artificial intelligence-based studies implementing computational machines with specific functions, such as automatic diagnosis and prognosis prediction for an individual patient, using machine learning techniques based on large amounts of data obtained from multiple patients and 2) patient-specific modeling-based studies implementing biophysical in-silico platforms to understand pathological mechanisms and derive the optimal treatment for each patient by reproducing the brain network dynamics of the particular patient per se based on individual patient’s data. These computational approaches are important as it can integrate multiple types of data acquired from patients and analysis results into a single platform. If these kinds of methods are efficiently operated, it would suggest a novel paradigm for precision medicine.
Introduction
Epilepsy is a chronic neurological disease that affects more than 50 million people worldwide.1 Epilepsy is defined as recurrence of unexpected seizures, which results in various ictal symptoms depending on the brain regions where the seizure is originated and propagated.2,3 It seriously degrades the quality of life not only of the individual patients, but of their family, which causes major socio-economic losses.4–6
For the treatment of epilepsy, medical therapy can be applied by priority, in which 60–70% of the patients have responded by single or multiple antiepileptic medications.7,8 For the drug-resistant patients, however, surgical intervention has been provided as an option, including resection that removes the focal area in the brain, which generates epileptic seizures, as well as disconnection that blocks the main neural pathways of seizure propagation.9–13 Although the epilepsy surgery has been accepted as an effective method to control the drug-resistant seizures, the postoperative outcomes have been largely variable depending on various clinical factors of the individual epilepsy patient, including etiology, diagnostic modalities and even decision makings by physicians or surgeons’ point of view.9–13 The possibility of cognitive impairment after surgery caused by the removal or severance of specific brain areas is another important issue to consider.14–17
Recently, neuromodulation therapy using various brain stimulation modalities, including deep brain stimulation, vagal nerve stimulation, or external responsive neurostimulation, have been attempted to control intractable seizures.18–22 Those therapies modulate brain functions or pathological states at the entire brain network level, by stimulating the specific brain region to induce changes not only in the stimulated brain regions but also in distant areas that are connected to them through the anatomical or functional brain connectivity. While the neuromodulation therapies have been known as novel and promising treatment methods, demonstrating seizure reduction in about one-third to one-half of patients,18–22 there are still many obstacles for improving the treatment efficacy by finding the optimal stimulation sites or stimulation parameters, or understanding the mechanisms behind the treatment.23,24
Although studies for the epilepsy treatment have been actively conducted in multiple avenues, one of the most difficult challenges is the between-subject variability of the treatment outcomes. In fact, the treatment outcomes have been varied from patient to patient depending on the patient-specific intrinsic characteristics, including seizure types and semiologies, brain lesions, or comorbid neuropsychological dysfunctions, extrinsic factors such as the stage in which the treatment is first applied, and the treatment conditions.25–28 Therefore, personalized approaches that diagnose each patient’s state accurately and choose an optimal treatment method to the individual patient are crucial.
With the advance of high-performance computing technologies and the development of numerous mathematical algorithms, computational studies for clinical application on epilepsy are being actively conducted to analyze large amounts of data, from which a proper solution can be derived for an individual patient (Fig. 1). These computational studies have significant impacts as it can provide automated and standardized protocols to support clinical decisions. This manuscript provides an overall introduction regarding recent computational studies on personalized medicine and discuss its future directions for diagnosis and treatment of epilepsy patients.
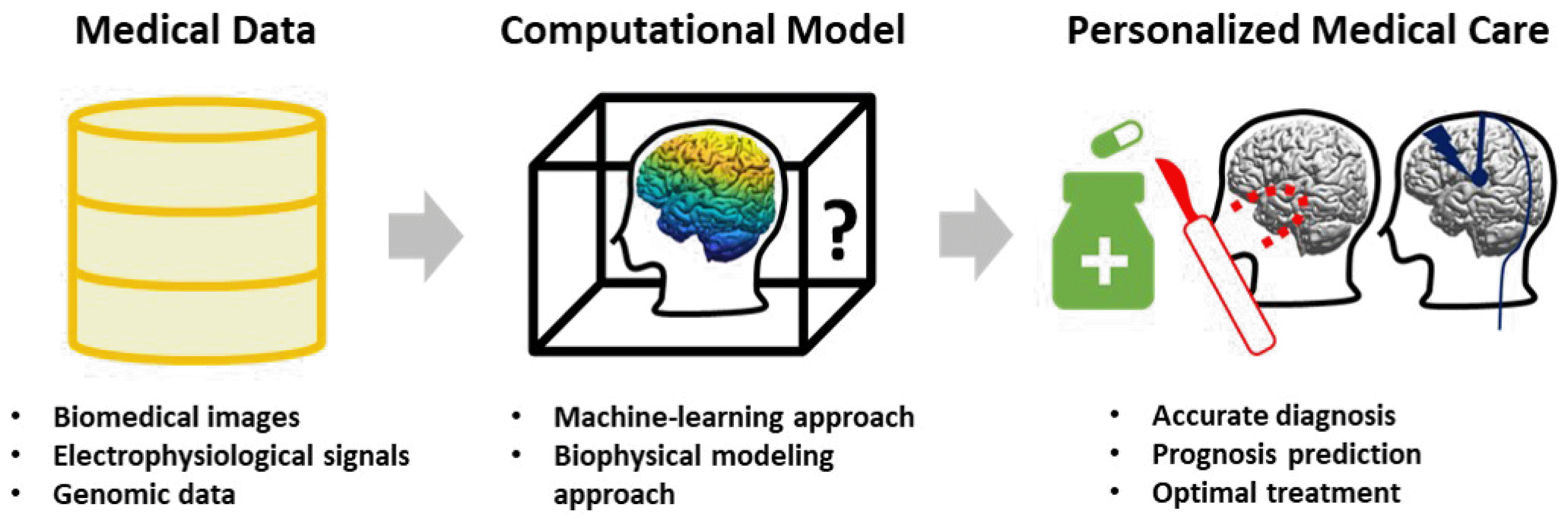
Overview of computational approaches for personalized medicine. Using accumulated large amount of medical data and advanced in-silico techniques, the personalized medical platform could be built to provide accurate diagnosis, prognosis prediction and treatment optimization for individual epilepsy patients. Currently, these computational studies are mainly divided into two approaches: a machine learning approach that implements a model capable of performing a specific function, such as automatic diagnosis, and a biophysical modeling approach that reproduces and simulates the patient’s brain network dynamics system itself.
Machine learning-based approaches
Machine learning, an application of artificial intelligence (AI) technique, enables a machine to automatically learn something new by combining statistics and computer science and thus improve its performance through meaningful data, without explicit instruction.29 These learning tasks are executed in two main types: supervised versus unsupervised learnings.29–33 Supervised learning is the approach that trains using labeled data, that is, data whose target outputs have already been known. It is mainly used for classification or regression purposes and its algorithm includes k-nearest neighbor (k-NN), linear/logistic regression, naïve Bayes, random forest, and support vector machine (SVM).29–33 On the other hand, unsupervised learning is the approach that trains using unlabeled data. It is mainly used for clustering or association analysis purposes and its algorithm includes k-means, k-medoids, fuzzy C-means, Gaussian mixture, hidden Markov model.29–33 Artificial neural network (ANN) is another machine learning algorithm that performs the learning task by mimicking the brain nervous system, including neuronal dynamics and synaptic plasticity, and is widely used in both supervised and unsupervised learning schemes.29,31,33–36 If there are more than two hidden layers constituting ANN, it is specifically called deep neural network (DNN), and using those models to achieve the learning function is called deep learning.33 Deep learning approach has a significant advantage in automatically discovering discriminative features from data and learning them compared to traditional machine learning approach, which requires an additional process to extract the features manually and apply them as inputs.31,33,36,37 Thus, recently, it has been aggressively used in various applications even if it needs higher computational power. For the implementation of deep learning in addition to general DNN model (i.e., multi-layer perceptron), various transformed models, such as convolutional neural network (CNN) specialized for analysis of image data and recurrent neural network (RNN) specialized for analysis of time-series data, have been actively employed.36,37 In this section, we briefly review some of the notable studies that have applied machine learning techniques to clinical application of epilepsy.
Classification of seizure and epilepsy types
Determining seizure types and epilepsy types of individual patients is the first step that should be performed to provide adequate treatment for each patient. Typically, these diagnostic procedures have been conducted by clinicians reviewing multiple forms of data obtained from each patient, including symptoms, etiologies, neuroimages, neurophysiological data such as electroencephalogram (EEG), and etc. Since not only are these tasks time-consuming and laborious, but also recent studies have indicated that it can often be difficult to differentiate the epilepsy types even by experienced clinicians,38 the automated models based on the standardized protocols become more and more important.
Recent studies have proposed the machine learning approaches that automatically execute those diagnostic tasks, especially classification of seizure types, and have evaluated their performance.39–45 Mainly, using scalp-EEG recordings labeled with seizure types (recorded from multiple patients), some studies have shown that the computational model that trained the spatiotemporal features of the specific seizure classes were able to classify the seizure types with a quite high accuracy.39–43 For the implementation of these models in addition to classic machine learning algorithms such as SVM and k-NN,39–40 deep learning algorithms such as CNN41–43 have been applied. Notably, Liu and colleagues41 have proposed a hybrid bilinear model that combines CNN and RNN. In their hybrid model, CNN and RNN extracted the spatial and temporal features of seizures recorded in scalp EEG, respectively, especially from short-time Fourier transform results of segments with a 1 second time window. Then, these two types of features were combined into second-order statistics through bilinear pooling. The proposed model has classified the seizure types effectively, with F1-scores of 97.4% and 97.2% in two datasets, containing 8 and 4 seizure classes, respectively.
Some other studies have built the computational models that categorize seizure types or epilepsy types by training using text-based data containing patients’ symptoms.44,45 Kassahun and colleagues45 have proposed the models that classifies two epilepsy types, temporal lobe epilepsy and extra-temporal lobe epilepsy, based on the ictal symptoms of each patient, using employing two machine learning methods, ontology-based and genetics-based algorithms, and the models have achieved 77.8% accuracy.
These machine learning-based classification systems can be used to quickly determine disease characteristics of individual patients in a standardized manner, and they can be further applied to suggest drug medications that are appropriate to each patient based on accumulated clinical evidences.
Localization of seizure onset zones (SOZ)
Investigating SOZ and propagation zones (PZ) is crucial to make accurate diagnosis and treatment plan for each epilepsy patient. Especially when surgical intervention is considered, the localization of SOZ is essential to determine the surgical resection margin, from which the area that can prevent occurrence of seizures by its removal and does not cause critical impairment to the normal brain functions (i.e., outside of eloquent areas) should be derived. Localization of the SOZ has been mainly conducted by measuring electrophysiological signals containing spontaneous seizures in an invasive manner, such as intracranial EEG (iEEG) or stereotactic EEG (sEEG), and then analyzing the recorded EEG signals.
Recently, several studies have presented the machine learning-based methodology to identify SOZ.46–53 In particular, using iEEG recordings from patients, Elahian and colleagues47 have built a model to classify each electrode position into SOZ and non-SOZ. They considered the electrode positions within the resected area and outside the resected area in the patients who showed seizure-free outcome as SOZ and non-SOZ, respectively, and trained the model using the signal characteristics recorded at the corresponding electrodes of two classes. For the training, they extracted certain features from the phase locking values (PLVs) of the signals for each channel, especially PLV between the phase of amplitude of high gamma activity and phase of lower frequency rhythms, and applied them to the logistic regression algorithm. They have demonstrated it in patients with poor postoperative outcomes, some of the SOZ electrodes predicted by the model remained outside the resected area, and the number of non-resected SOZ electrodes correlated with surgical outcomes.
In contrast to the studies that localize SOZ using the recording data containing spontaneous seizures47–49 as described above, other studies have attempted to perform the same task using only recording data during inter-ictal period.50–53 Varatharajah and colleagues51 have developed an analytic framework to make localization of SOZ possible, based on multiple biomarkers analyzed from the inter-ictal iEEG recordings, including high frequency oscillation, interictal epileptiform discharge, and phase amplitude coupling. The model derived by training the features extracted from the three biomarkers through the SVM algorithm was able to effectively identify the SOZ using 2 hours of interictal recordings (with an average area under ROC curve value of 0.73, when compared to clinically investigated SOZ).
With further validation, these machine learning-based SOZ localization systems can be used to assist decision-making process in determining the surgical site when surgical intervention is considered in drug-resistant epilepsy patients (Fig. 2).
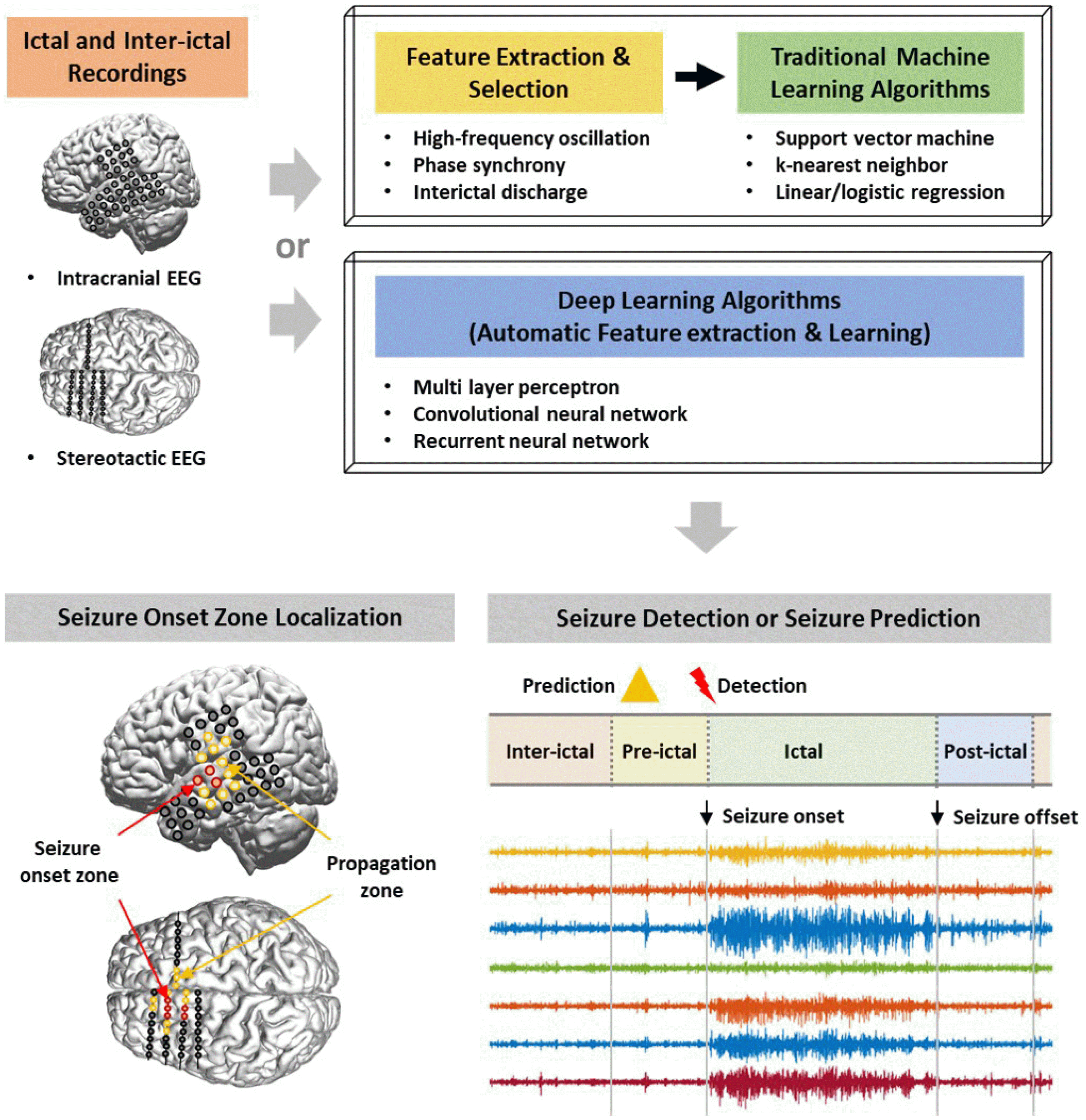
Machine learning-based computational approach. Many recent studies have focused on implementing the computational predictive models to localize SOZs or judge epileptic brain states, such as pre-ictal and ictal onset, by employing traditional machine learning algorithms or deep learning algorithms based on scalp EEG and/or iEEG data recorded from epilepsy patients. Compared to the traditional machine learning approach, which consists of two step processes of manually extracting the features of the data and training the machine by applying the features as inputs, deep learning approach automatically figures out the discriminative features from data and learns them. SOZ, seizure onset zones; EEG, electroencephalogram; iEEG, intracranial EEG.
Examination of epileptic brain states
Epileptic brain dynamics can be divided into four states: inter-ictal (period between seizures, i.e., a normal state of patients), pre-ictal (period immediately before the seizure onset), ictal (period during seizure), and post-ictal (period immediately after the seizure).54 Investigating the characteristics of each of these states and the transitions between them is significant not only for understanding the pathological mechanisms of the epilepsy, but also treatment and disease management. To date, these studies have been conducted with two main purposes: seizure detection that promptly identifies the seizure onset (time-wise) and seizure prediction that forecast the occurrence of seizure in advance by recognizing the characteristics of the pre-ictal state different from inter-ictal. Recently, numerous studies have proposed computational models that can automatically perform these tasks using accumulated data sets and machine learning algorithms.
Regarding the seizure detection task, many studies have shown that the models trained via traditional machine learning algorithms,55–60 especially SVM and k-NN, deep learning algorithms61–65 using specific features in time and/or frequency domain based on scalp EEG, or iEEG recordings can successfully detect seizures with multiple types. In particular, Emami and colleagues have proposed a CNN-based seizure detection model.61 The model learned the EEG (scalp EEG) characteristics in seizure state and non-seizure state automatically, without additional manual feature extraction procedures, through the supervised learning framework, and was able to detect seizure onset at an average positive rate of 74% when the entire time series EEG was sequentially input by 1 second (100% for input by 1 minute). They also have demonstrated that performance of seizure detection depends on the similarity of seizure onset pattern between training data and test data, i.e., new data with a different onset pattern that those of trained data could not be detected well. These results indicated that in order to achieve the model that conducts seizure detection with high performance, it is necessary to train using a large amount of data including various seizure patterns. These computational models with the purpose of seizure onset detection can be used to provide basis for on-demand (closed-loop) stimulation therapies or acute drug treatment.
In relation to the seizure prediction task, numerous studies have reported that the computational models that learned time and/or frequency domain features observed in pre-ictal state was able to predict the occurrence of seizures at least several minutes before the onset. These models have been mainly trained through supervised learning methods, and they were based on a variety of algorithms ranging from classic machine learning algorithms such as SVM,66–71 k-NN,71–73 hidden Markov model,74 and etc., to deep learning algorithms such as CNN,75–78 Long Short-Term Memory70,79 (LSTM, a kind of RNN) and their hybrid model,80–82 learning the characteristics of the pre-ictal state distinct from inter-ictal. The developed models have shown sensitivity of 80–90%, but it should be noted that each study had a different prediction time (from 5 minutes before to 1 hour before the onset). Currently, studies of seizure prediction have been conducted primarily in the direction of developing patient-specific models based on data obtained from individual patients, rather than developing a generalized model based on lots of data from multiple patients. Given that the signal patterns of iEEG or scalp EEG vary for each patient, this individualized approach may be more useful for clinical applications. Recent studies have implemented seizure prediction models mainly by employing deep learning algorithms.75–82 Notably, Daoud and Bayoumi80 have developed a model that predicts seizures 1 hour before the onset, with a high accuracy of 99.6%, using long-term scalp EEG. The proposed model employed both CNN and RNN (especially bidirectional LSTM) to learn the spatial and temporal features respectively from raw EEG data and introduced a semi-supervised learning approach based on the transfer learning technique to reduce training time, showing the potential for real-time usage. Meanwhile, Cook and colleagues have demonstrated the feasibility of the implanted seizure advisory system in drug-resistant patients,83 in which they implemented a machine learning-based patient-specific algorithm by using iEEG signals recorded for at least 1 month (containing at least 5 leading seizures) at each patient and predicted seizure likelihood based on that algorithm, achieving sensitivities ranging from 65% to 100% for each patient. Moreover, Kiral-Kornek and colleagues84 have demonstrated feasibility of real-time using wearable devices with low power consumption and long-term reliability by implementing the patient-specific seizure prediction system onto neuromorphic chip. These machine learning-based seizure prediction systems not only help the patients to avoid dangerous situations by alerting them of the likelihood of seizures, but also establish the basis of offering personalized treatment by providing each patient’s information including frequency, duration, and patterns of seizures to medical institutions (Fig. 2).
Biophysical modeling-based approaches
As another branch of the computational approaches that is distinct from machine learning-based approach, which create the computational machine to performing specific functions (especially, such as classification or clustering) inspired by the working mechanisms of the neural network, computational modeling approach that reproduces the neural network dynamics per se is also actively underway.85–88 These neural network modeling studies are mainly performed in two types of approaches: bottom-up approach of constructing a network based on microscale units including single neurons, synapses and ion channels, and top-down approach of building a whole-brain network model based on macroscale brain con-nectome to understand its functions and mechanisms. In particular, since a framework to construct the subject-specific brain network model based on individual neuroimaging data has been developed,89–90 brain modeling studies of the top-down approach to investigate brain functions and dysfunctions in a personalized manner have been rapidly progressing. In these models at whole brain level, neural mass model, which is a mathematical model that describes the activities of neuronal population rather than the activities of single neurons, is mainly used. These neural mass models are located in each brain region (or, in each sensor location), generating local dynamics, and interact each other by being connected via brain connectivity acquired from the individual brain imaging data. The parameters of each neural mass model that determine the local regional properties can be set through the analysis of individual functional imaging data or through the clinical findings. This personalized modeling approach can be applied to specific diseases, and the developed model could reproduce the pathological characteristics of each patient, such as structural and/or functional alteration of the brain,17,91–93 and predict the effects of various therapeutic interventions via systematic simulations in a patient-specific environment. In this section, we briefly review the personalized modeling studies toward clinical application of epilepsy.
Patient-specific whole-brain models
To date, studies of personalized brain network modeling have been primarily aimed at optimizing surgical intervention strategies by employing retrospective approach, in which the actual surgical site of each patient is compared with the target site derived from the simulations using the model, and those comparison results are analyzed with the surgical outcomes. These patient-specific models have been constructed in two main ways: sensor-based and region-based. In the sensor-based models, neural mass models are positioned in each sensor location, i.e., electrode position of iEEG or scalp EEG, and they are coupled by the functional connectivity analyzed from the corresponding data.94–96 In the region-based models, neural mass models are located in each brain region and they are coupled by structural brain connectivity analyzed from structural brain imaging data, mainly T1-weighted and diffusion-weighted images.97–102 The retrospective modeling studies have demonstrated that poor surgical outcomes are frequently observed when target sites identified by simulations are not sufficiently resected during actual surgery,96,99 In other words, they indicate that in-depth analysis considering network dynamics based on the data is required rather than interpretation of the data itself in order to determine the optimal surgical target.
Jirsa’s group has been intensively conducting personalized brain network modeling studies for clinical application, mainly adopting region-based modeling scheme. They have proposed a novel methodology to develop individual brain network models, so called the Virtual Epileptic Patient (VEP),98 by incorporating multimodal data from each epilepsy patient, and have also developed and distributed the neuroinformatics platform, The Virtual Brain, to enable the model construction.89–90 The VEP model was constructed based on Epileptor,103,104 a phenomenological neural mass model replicating seizure characteristics, and patient-specific features including brain connectivity and magnetic resonance imaging lesions, and fit and validated using the individual sEEG recordings.98,99 Based on this personalized modeling approach, Jirsa’s group has been conducting successive studies in various directions for clinical applications, such as inference of the epileptogenicity of the brain regions,102 analysis of the seizure propagation characteristics,99–101 and prognostic prediction of the surgical Intervention.100,101
Proix et al.99 have created the personalized brain network models for 15 epilepsy patients and simulated individual seizure propagation patterns. They also have explained the variability of surgical outcomes using the simulated seizure propagation patterns, demonstrating that the number of regions, which were identified as the PZ in the simulation but not considered in the pre-surgical evaluation (because they were not investigated by sEEG), were correlated with poor surgical outcomes. Some other researchers have performed simulations to derive the surgical target sites that are minimally invasive and effective in suppressing seizures, by applying an in-silico surgical approach to the personalized model, i.e., by investigating the degree of seizure propagation in pre- and post-surgical conditions.100,101 An et al.101 have proposed a strategy to evaluate the safety of the target sites in order not to cause impairment of normal brain functions, in which they operationalized the safety by the concept of preservation of signal transmission capacity of the brain network, by comparing the simulation-induced response network characteristics in pre- and post-surgical conditions. These research outcomes have shown the possibility that the personalized brain network model could support clinical decision in determining the target site for surgery, and Jirsa’s group are currently applying this modeling approach to large-scale clinical trials in order to evaluate its effectiveness.
These personalized network modeling approaches can be applied to investigate the effect of various brain interventions such as neuro-stimulation in addition to surgical treatment and thus can be utilized to understand the therapeutic mechanisms and suggest optimal treatment method for each patient (Fig. 3).

Biophysical modeling-based computational approach. Recent studies based on personalized brain network modeling approach have demonstrated the its feasibility, in which the model can reproduce seizure propagation characteristics of each patient and suggest optimal intervention method for each patient via systematic simulations in the patient-specific environments.
Conclusions
In this paper, we have reviewed the computational studies that have been conducted for clinical applications in epilepsy, especially in terms of AI-based approach to generate a computational machine performing specific functions, and biophysical modeling-based approach to replicate neural network dynamics per se. Numerous studies have demonstrated the feasibility of computational approaches that could build a novel medical paradigm with a personalized manner, encompassing diagnosis, prognosis and optimization of treatment, by integrating multimodal data and their analytical results into a single platform. For successful clinical application of the developed computational systems and commercialization through optimization, close interdisciplinary cooperation in various fields including medicine, neuroscience, computer science, and engineering is crucial.
Acknowledgements
This study was supported by grants from the Basic Science Research Program and Convergent Technology R&D Program for Human Augmentation through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, Information and Communication Technologies & Future Planning (2017R1A2A2A050696, 2017R1B5A2086553, 2018M3C1B8016147, 2019M3C1B8090803, and 2020R1A2C2013216) to H. W. Lee. This research was also supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1I1A1A01073605) to S. An.
Notes
Conflict of Interest
The authors declare that they have no conflicts of interest.